TP3 : Lazy training#
Observing the lazy training regime with different models
With inspiration from
https://rajatvd.github.io/NTK/
import torch
import torch.optim as optim
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
import matplotlib.pyplot as plt
### Exectutes the script tp3_utils.py which contains code from last time
## Go take a quick look at the functions implemented there
from tp3_utils import *
# %load_ext autoreload
# %autoreload 2
The data#
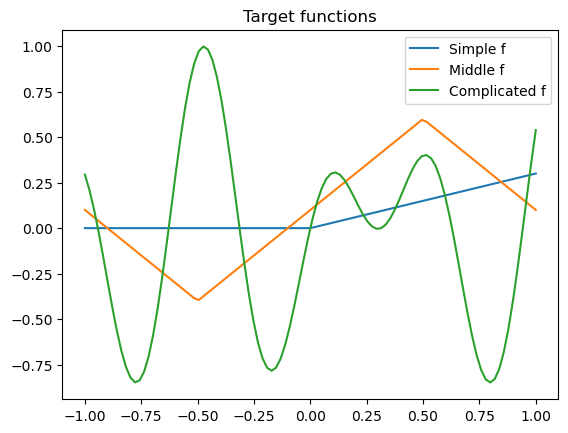
We continue with last week’s toy problem of 1 dimensional regression
xs = np.linspace(-1, 1, 100)
plt.plot(xs, simple_f(xs), label="Simple f")
plt.plot(xs, middle_f(xs), label="Middle f")
plt.plot(xs, complex_f(xs), label="Complicated f")
plt.title("Target functions")
plt.legend()
plt.show()
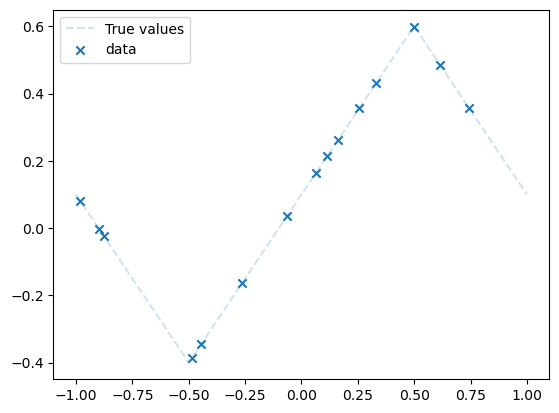
dataset = Data(n=15, xmin=-1, xmax=1, noise_level=0, type="middle")
plot_data(dataset)
plt.legend()
plt.show()
The training function#
Write the training function
def train(net, dataset, N_steps=1, batch_size=30, lr=0.01, save_weights_every=100):
optimizer = optim.SGD(net.parameters(), lr=lr)
criterion = nn.MSELoss()
losses = []
weights = []
for i in range(N_steps):
inputs, labels = dataset.next_batch(batch_size)
optimizer.zero_grad()
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
losses.append(loss.detach().numpy())
if i % save_weights_every == 0 and (save_weights_every > 0):
with torch.no_grad():
weights.append(net.save_weights())
return losses
def train(net, dataset, N_steps=1, batch_size=30, lr=0.01, save_weights_every=100):
# YOUR CODE HERE
losses = []
weights = []
for i in range(N_steps):
# YOUR CODE HERE
losses.append(loss.detach().numpy())
if i % save_weights_every == 0 and (save_weights_every > 0):
with torch.no_grad():
weights.append(net.save_weights())
return losses
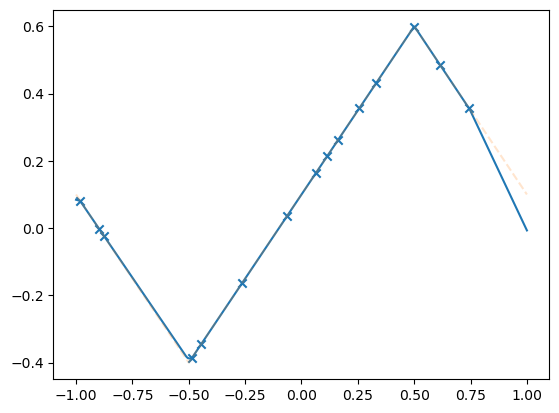
Test your training#
model = MLPdeep(hidden_dim=10)
losses = train(
model,
dataset,
N_steps=10000,
batch_size=15,
lr=0.1,
save_weights_every=1,
)
plot_net(model)
plot_data(dataset)
plt.show()
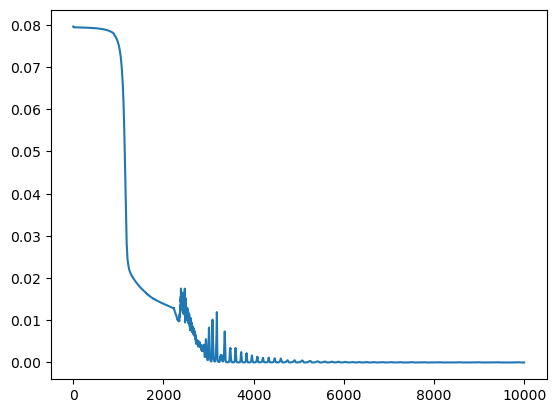
plt.plot(losses)
plt.show()
A linear model#
We define a family of linear models with sinusoidal features. The feature map is
where \(k\) is feature_dim / 2.
class LinSin(nn.Module):
"""
A linear model with sinusoidal features. Assumes feature_dim is even.
"""
def __init__(self, feature_dim):
super().__init__()
self.feature_dim = feature_dim
self.lout = nn.Linear(self.feature_dim, 1)
self.weight_history = []
def phi(self, x):
"""
Computes $sin(2\pi k x), cos(2\pi k x) $ for k in $[feature_dim / 2]$
"""
r = 2 * np.pi * torch.arange(int(self.feature_dim / 2)) * x
return torch.concatenate([torch.sin(r), torch.cos(r)], axis=1)
def save_weights(self):
with torch.no_grad():
l = list(self.parameters())
self.weight_history.append(np.copy(l[-1].numpy()))
def forward(self, x):
return self.lout(self.phi(x))
class LinSin(nn.Module):
"""
A linear model with sinusoidal features
"""
def __init__(self, feature_dim):
super().__init__()
self.feature_dim = feature_dim
self.lout = nn.Linear(self.feature_dim, 1)
self.weight_history = []
def phi(self, x):
"""
Computes $sin(2\pi k x), cos(2\pi k x) $ for k in $[feature_dim / 2]$
"""
# YOUR CODE HERE
pass
def save_weights(self):
with torch.no_grad():
l = list(self.parameters())
self.weight_history.append(np.copy(l[-1].numpy()))
def forward(self, x):
# YOUR CODE HERE
pass
Test the linear model
model = LinSin(feature_dim=8)
losses = train(
model,
dataset,
N_steps=10000,
batch_size=15,
lr=0.01,
save_weights_every=1,
)
plot_net(model)
plot_data(dataset)
plt.show()
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].loglog(losses)
axs[0].set_title("Train loss")
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(all_weights1 - all_weights1[0], axis=1) / np.linalg.norm(
all_weights1[0]
)
axs[1].plot(weight_evol, alpha=0.5)
axs[1].set_title("Relative weight movement of last layer")
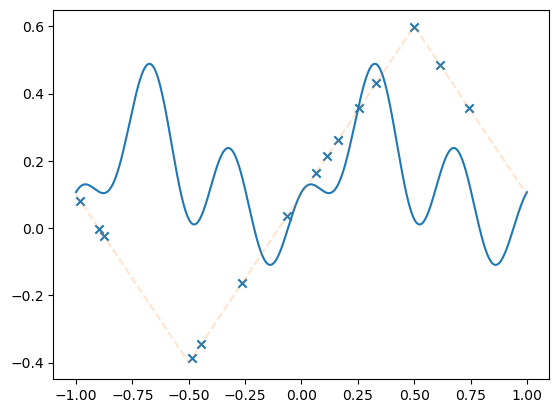
Text(0.5, 1.0, 'Relative weight movement of last layer')
Question: what do the plots mean?
Let us now repeat the code above with a large feature dimension. What you do observe?
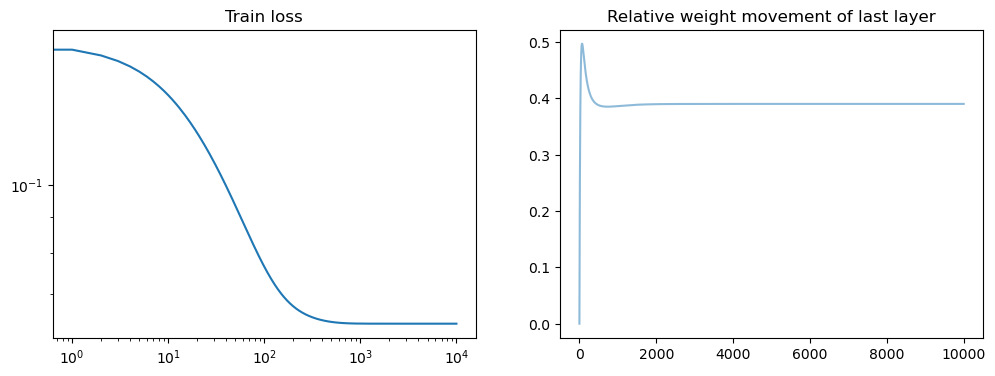
model = LinSin(feature_dim=200)
losses = train(
model,
dataset,
N_steps=100,
batch_size=15,
lr=0.01,
save_weights_every=1,
)
plot_net(model)
plot_data(dataset)
plt.title("Trained model")
plt.show()
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].loglog(losses)
axs[0].set_title("Train loss")
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(all_weights1 - all_weights1[0], axis=1) / np.linalg.norm(
all_weights1[0]
)
axs[1].plot(weight_evol, alpha=0.5)
axs[1].set_title("Relative weight movement of last layer")
plt.show()
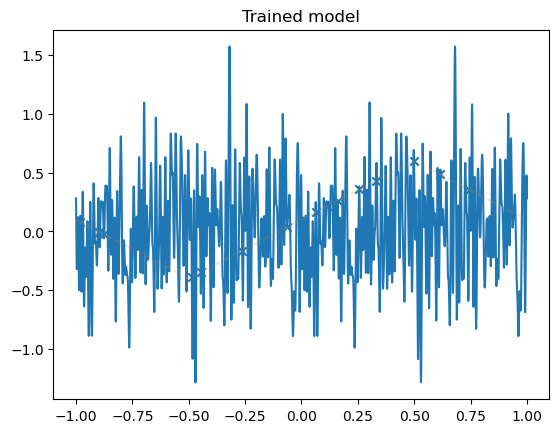
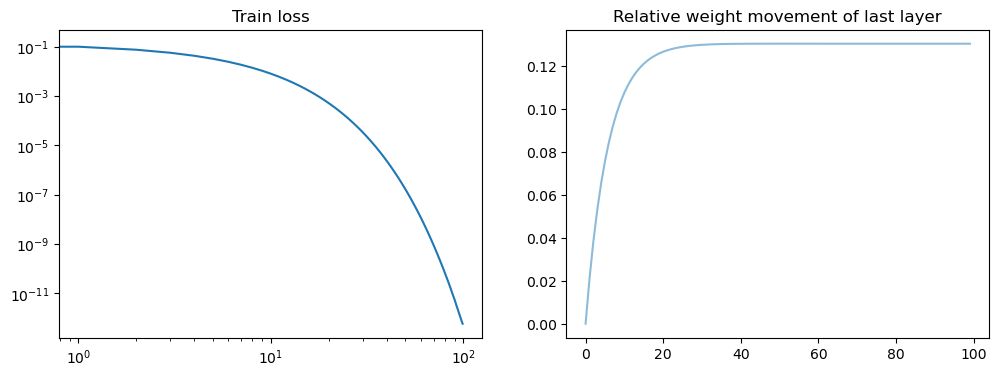
Robustness of fast convergence for linear models#
Run 10 instances of the linear model with different initializations.
Are there significant differences between the models?
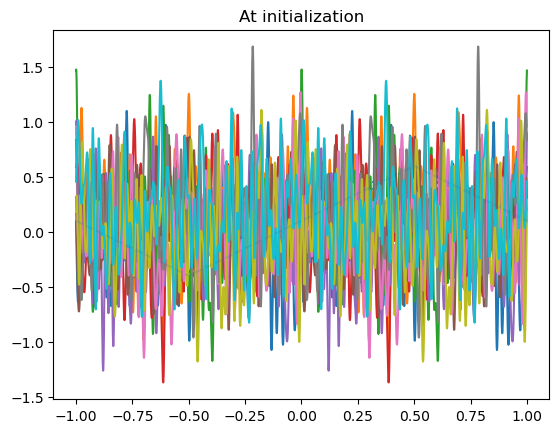
model_list = []
N_models = 10
for _ in range(N_models):
model = LinSin(feature_dim=100)
model_list.append(model)
plot_net(model)
plot_data(dataset)
plt.title("At initialization")
all_losses = [[] for _ in range(N_models)]
all_weights = []
N_steps = 10000
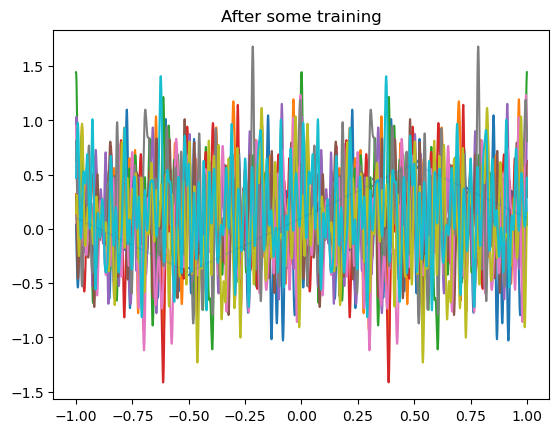
for i, model in enumerate(model_list):
losses = train(
model,
dataset,
N_steps=N_steps,
batch_size=15,
lr=0.01,
save_weights_every=1,
)
all_losses[i] += losses
plot_net(model)
plot_data(dataset)
plt.title("After some training")
plt.show()
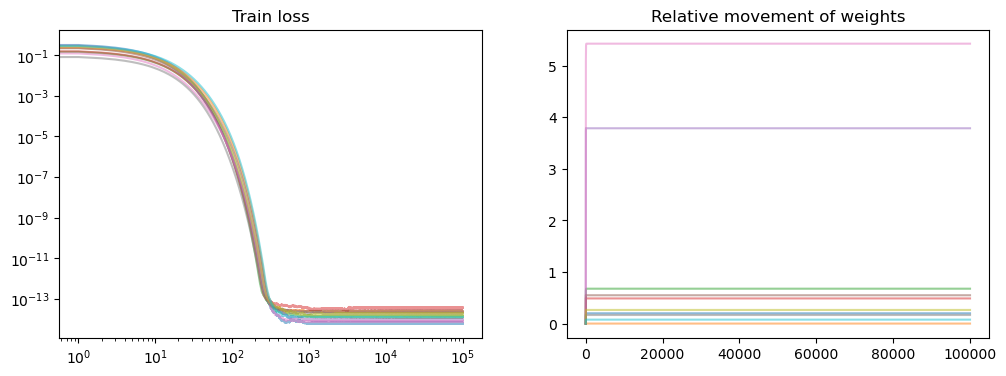
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].set_title("Train loss")
for losses in all_losses:
points = np.arange(0, len(losses), 1)
losses = np.array(losses)
axs[0].loglog(points, losses[points], alpha=0.5)
axs[1].set_title("Relative movement of weights")
for model in model_list:
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(
all_weights1 - all_weights1[0], axis=1
) / np.linalg.norm(all_weights1[0])
axs[1].plot(weight_evol, alpha=0.5)
plt.show()
Lazy training of non-linear models#
We now train some neural nets in different regimes and try to see when lazy training occurs.
Question How are nets initialized in pytorch?
https://pytorch.org/docs/stable/generated/torch.nn.Linear.html
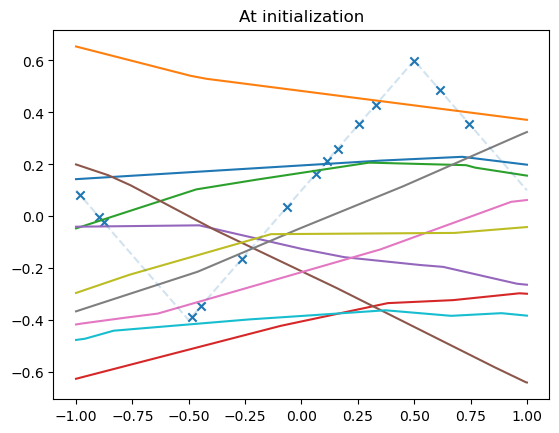
model_list = []
N_models = 10
for _ in range(N_models):
model = MLPshallow(hidden_dim=10)
model_list.append(model)
plot_net(model)
plot_data(dataset)
plt.title("At initialization")
all_losses = [[] for _ in range(N_models)]
all_weights = []
print(sum([p.numel() for p in model.parameters()]))
31
N_steps = 5000
for i, model in enumerate(model_list):
losses = train(
model,
dataset,
N_steps=N_steps,
batch_size=30,
lr=0.001,
save_weights_every=1,
)
all_losses[i] += losses
plot_net(model)
plot_data(dataset)
plt.title("After some training")
plt.show()
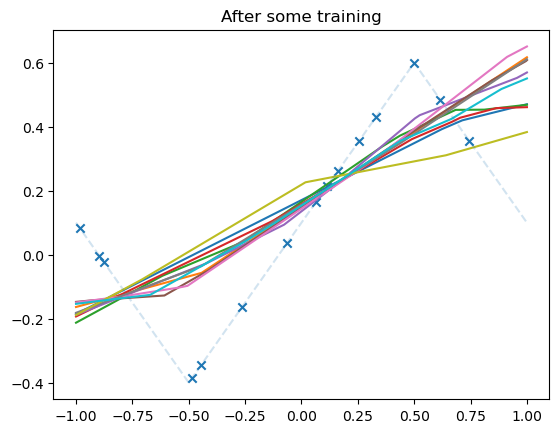
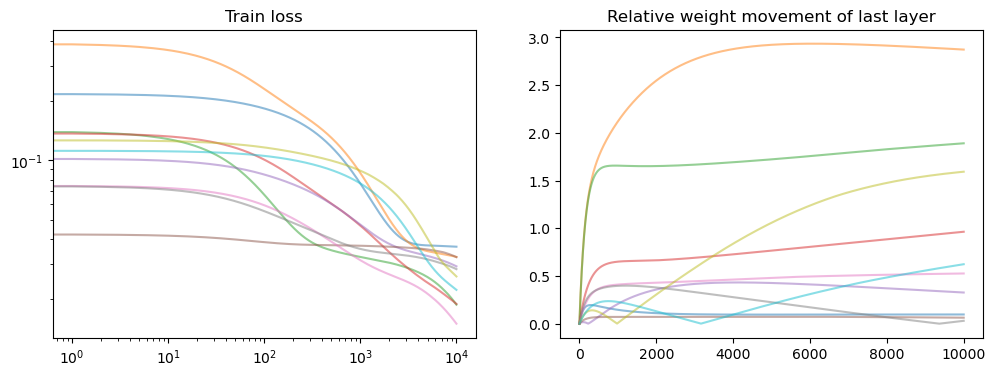
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].set_title("Train loss")
for losses in all_losses:
points = np.arange(0, len(losses), 1)
losses = np.array(losses)
axs[0].loglog(points, losses[points], alpha=0.5)
axs[1].set_title("Relative weight movement of last layer")
for model in model_list:
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(
all_weights1 - all_weights1[0], axis=1
) / np.linalg.norm(all_weights1[0])
axs[1].plot(weight_evol, alpha=0.5)
plt.show()
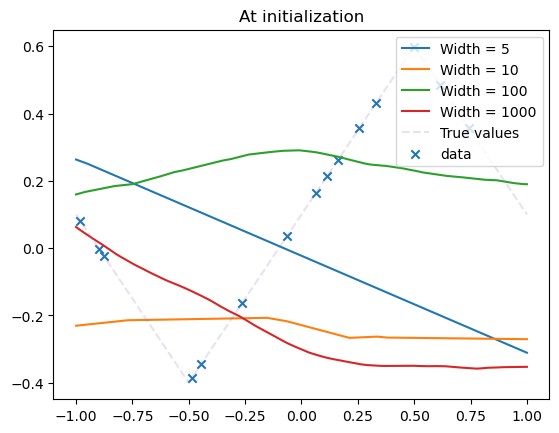
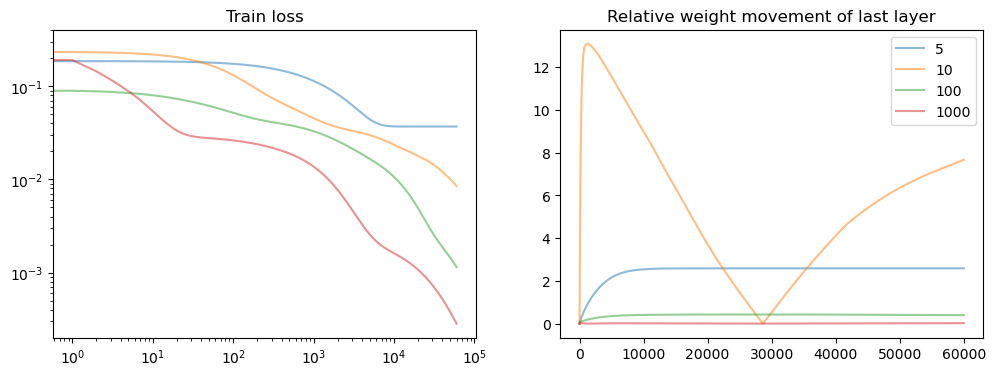
Thin vs. Wide networks#
Let us now vary the width of our network. What happens?
model_list = []
width_list = [5, 10, 100, 1000]
for width in width_list:
model = MLPshallow(hidden_dim=width)
model_list.append(model)
plot_net(model, label=f"Width = {model.hidden_dim}")
plot_data(dataset)
plt.title("At initialization")
plt.legend()
all_losses = [[] for _ in width_list]
all_weights = []
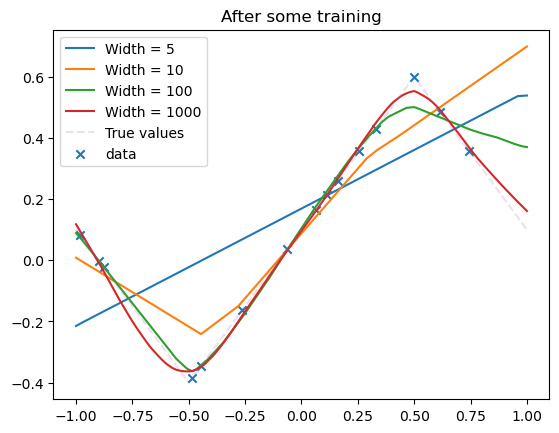
N_steps = 50000
for i, model in enumerate(model_list):
losses = train(
model,
dataset,
N_steps=N_steps,
batch_size=15,
lr=0.002,
save_weights_every=1,
)
all_losses[i] += losses
plot_net(model, label=f"Width = {model.hidden_dim}")
plot_data(dataset)
plt.title("After some training")
plt.legend()
plt.show()
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].set_title("Train loss")
for i, losses in enumerate(all_losses):
points = np.arange(0, len(losses), 1)
losses = np.array(losses)
axs[0].loglog(points, losses[points], alpha=0.5, label=f"{width_list[i]}")
axs[1].set_title("Relative weight movement of last layer")
for model in model_list:
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(
all_weights1 - all_weights1[0], axis=1
) / np.linalg.norm(all_weights1[0])
axs[1].plot(weight_evol, alpha=0.5, label=f"{model.hidden_dim}")
plt.legend()
plt.show()
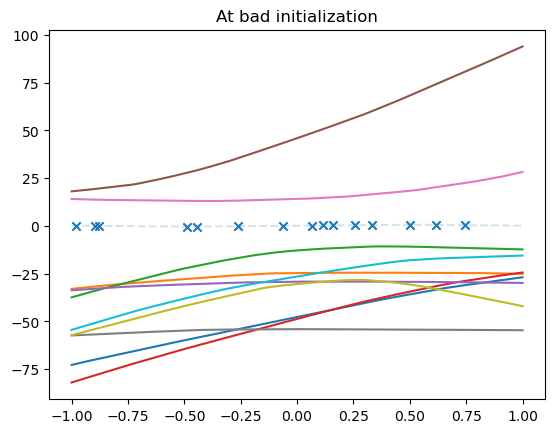
Bad initialization can you get you out of the Lazy regime#
We simulate bad initialization by taking two steps of gradient descent from initialization.
model_list = []
N_models = 10
for _ in range(N_models):
model = MLPshallow(hidden_dim=100)
model_list.append(model)
train(
model,
dataset,
N_steps=2,
batch_size=30,
lr=1,
save_weights_every=-1,
)
plot_net(model)
plot_data(dataset)
plt.title("At bad initialization")
all_losses = [[] for _ in range(N_models)]
all_weights = []
N_steps = 1000
xmin = np.min(dataset.inputs.detach().numpy())
xmax = np.max(dataset.inputs.detach().numpy())
plt.xlim(xmin, xmax)
print(xmin, xmax)
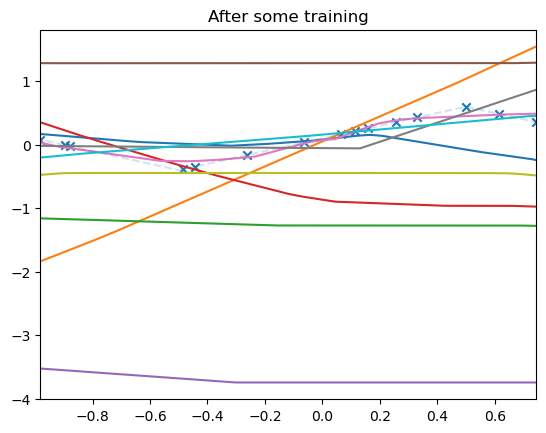
for i, model in enumerate(model_list):
losses = train(
model,
dataset,
N_steps=N_steps,
batch_size=30,
lr=0.001,
save_weights_every=1,
)
all_losses[i] += losses
plot_net(model, xmin=xmin, xmax=xmax)
plot_data(dataset)
plt.title("After some training")
plt.show()
-0.98198813 0.7443142
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].set_title("Train loss")
for i, losses in enumerate(all_losses):
points = np.arange(0, len(losses), 1)
losses = np.array(losses)
axs[0].loglog(points, losses[points], alpha=0.5)
axs[1].set_title("Relative weight movement of last layer")
for model in model_list:
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(
all_weights1 - all_weights1[0], axis=1
) / np.linalg.norm(all_weights1[0])
axs[1].plot(weight_evol, alpha=0.5)
plt.show()
Scaling#
Now we attempt to enter the lazy regime by scaling the model outputs.
n_points = 10
dataset = Data(n=n_points, xmin=-1, xmax=1, noise_level=0, type="middle")
class ScaledModel(MLPdeep):
""" "
From a base network, creates a scaled copy with 0 initialization. Keeps a frozen copy of a base model.
alpha: scaling factor
"""
def __init__(self, base_model, alpha=1):
super().__init__(hidden_dim=base_model.hidden_dim)
self.alpha = alpha
self.load_state_dict(base_model.state_dict())
self.base_model = [
base_model
] # trick to hide from the other parameters dictionary. Probably not the proper way to do this...
def forward(self, x):
"""
Scales the output and subtracts the initial function
"""
with torch.no_grad():
base = self.base_model[0].forward(x).detach()
return self.alpha * (super().forward(x) - base)
base_model = MLPdeep(hidden_dim=8)
alphas = [1, 100, 1000] # , 2000, 10000]
model_list = []
for alpha in alphas:
new_model = ScaledModel(base_model, alpha=alpha)
new_model.load_state_dict(base_model.state_dict())
model_list.append(new_model)
plot_net(new_model, label=f"{alpha}")
all_losses = [[] for _ in range(len(model_list))]
plt.legend()
plt.show()
train(
model_list[2],
dataset,
N_steps=10000,
batch_size=n_points,
lr=0.1 / alphas[2] ** 2,
save_weights_every=1,
)
plot_data(dataset)
plt.legend()
plt.title("After some training")
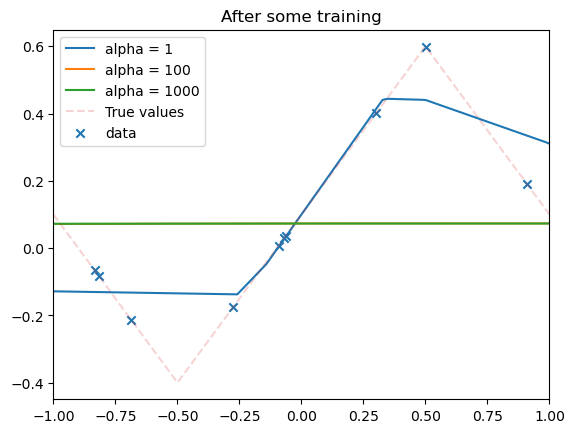
for i, model in enumerate(model_list):
plot_net(model, xmin=xmin, xmax=xmax, label=f"alpha = {alphas[i]}")
N_steps = 10000
xmin = -1 # np.min(dataset.inputs.detach().numpy())
xmax = 1 # np.max(dataset.inputs.detach().numpy())
plt.xlim(xmin, xmax)
print(xmin, xmax)
for i, model in enumerate(model_list):
losses = train(
model,
dataset,
N_steps=N_steps,
batch_size=n_points,
lr=0.01 / alphas[i] ** 2,
save_weights_every=1,
)
all_losses[i] += losses
plot_net(model, xmin=xmin, xmax=xmax, label=f"alpha = {alphas[i]}")
plot_data(dataset)
plt.legend()
plt.title("After some training")
plt.show()
-1 1
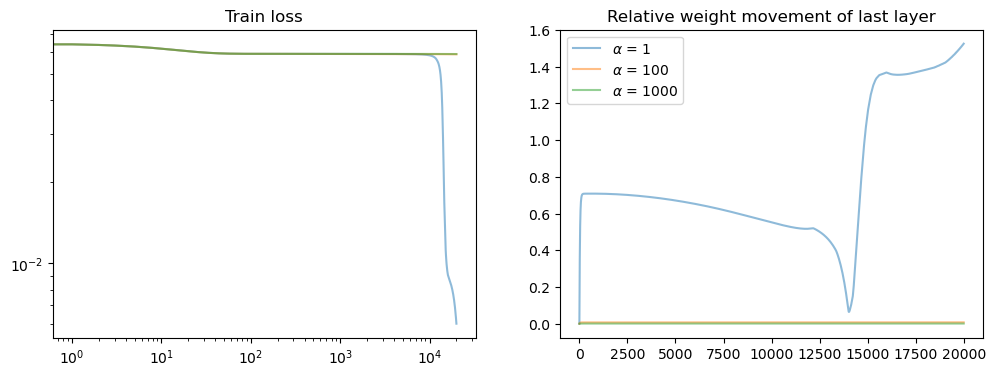
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].set_title("Train loss")
for i, losses in enumerate(all_losses):
points = np.arange(0, len(losses), 1)
losses = np.array(losses) # / alphas[i] ** 2
axs[0].loglog(points, losses[points], alpha=0.5, label=f"{alphas[i]}")
axs[1].set_title("Relative weight movement of last layer")
for i, model in enumerate(model_list):
all_weights1 = np.array(model.weight_history)
weight_evol = np.linalg.norm(
all_weights1 - all_weights1[0], axis=1
) / np.linalg.norm(all_weights1[0])
axs[1].plot(weight_evol, alpha=0.5, label=rf"$\alpha$ = {alphas[i]}")
plt.legend()
plt.show()
5 - The effect of scaling on a 1D model#
Question Explain the piece of code below.
see blog post
import numpy as np
import matplotlib.pyplot as plt
w_0 = 1.5
def f(x, w):
return (
w * x / 2 + np.log(1 + w**2) * x / 5 + 0.1 * np.sin(np.exp(2 * w))
) # np.exp(0.1*w))
def h(x, w, w_0=0.4):
return f(x, w) * (w - w_0)
def linh(x, w, w_0=0.4):
return f(x, w_0) * (w - w_0)
xys = [(1.5, 3)]
def l(w, alpha):
return np.sum([(alpha * h(x, w, w_0=w_0) - y) ** 2 / 2 for x, y in xys]) / alpha**2
def linl(w, alpha):
return (
np.sum([(alpha * linh(x, w, w_0=w_0) - y) ** 2 / 2 for x, y in xys]) / alpha**2
)
alphas = [1, 5, 10, 50, 100, 1000, 2000, 10000]
ws = np.linspace(-1, 1, 100)
plt.title("h(2; w)")
plt.plot(ws, [h(2, w, w_0=w_0) for w in ws])
plt.show()
for alpha in alphas:
delta = 2 / alpha ** (3 / 4)
ws = np.linspace(w_0 - delta, w_0 + delta, 5000)
plt.title(f"Loss landscape of scaled model: alpha = {alpha}")
plt.plot(ws, [l(w, alpha) for w in ws], label="Loss of scaled model")
plt.plot(ws, [linl(w, alpha) for w in ws], label="Loss of scaled linearized model")
plt.grid(alpha=0.2)
plt.legend()
plt.show()
import numpy as np
import matplotlib.pyplot as plt
w_0 = 1.5
def f(x, w):
return w * x / 2 + np.log(1 + w**2) * x / 5 + 0.1 * np.sin(np.exp(2 * w))
def h(x, w, w_0=0.4):
return f(x, w) * (w - w_0)
def linh(x, w, w_0=0.4):
return f(x, w_0) * (w - w_0)
xys = [(1.5, 3)]
def l(w, alpha):
return np.sum([(alpha * h(x, w, w_0=w_0) - y) ** 2 / 2 for x, y in xys]) / alpha**2
def linl(w, alpha):
return (
np.sum([(alpha * linh(x, w, w_0=w_0) - y) ** 2 / 2 for x, y in xys]) / alpha**2
)
alphas = [1, 5, 10, 50, 100, 1000, 2000, 10000]
for alpha in alphas:
delta = 2 / alpha ** (3 / 4)
ws = np.linspace(w_0 - delta, w_0 + delta, 5000)
plt.title(f"??: alpha = {alpha}")
plt.plot(ws, [l(w, alpha) for w in ws], label="??")
plt.plot(ws, [linl(w, alpha) for w in ws], label="??")
plt.grid(alpha=0.2)
plt.legend()
plt.show()
Pieces#
mlp = model_list[0]
for name, param in mlp.named_parameters():
print(name)
print(param.shape)
print(np.linalg.norm((param.detach().numpy())))
a = np.arange(10, 30)
shifted_a = np.zeros(20)
print(shifted_a)
shifted_a[1:] = a[:-1]
print(shifted_a)
print(a)
evol = (a - shifted_a)[1:] / a[1:]
print(evol)
plt.title("Difference in succesive weights")
for model in model_list:
all_weights1 = np.array(model.weight_history)
shifted_weights = np.zeros(all_weights1.shape)
shifted_weights[1:, :] = all_weights1[:-1, :]
weight_evol = np.linalg.norm(
(all_weights1 - shifted_weights)[1:], axis=1
) / np.linalg.norm(all_weights1[1:], axis=1)
plt.plot(weight_evol, alpha=0.5)
plt.show()
You can use the following code snippet to save your network.
# mlp = MLPdeep() # MLPshallow()
# PATH = f'./{dataset.type}_{mlp.net_type}.pth'
# torch.save(mlp.state_dict(), PATH)
# net = MLPdeep() # MLPshallow()
# net.load_state_dict(torch.load(PATH))
p = 100
n = 5000
X = np.random.normal(size=(p, n)) / np.sqrt(n)
Y = np.random.normal(size=(n, 1))
XXt = X @ X.transpose()
XY = X @ Y
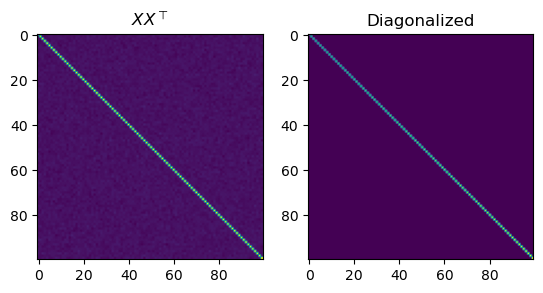
fig, axs = plt.subplots(1, 2)
axs[0].set_title(r"$X X^\top $")
axs[0].imshow(XXt)
eigvals, P = np.linalg.eigh(XXt)
print(max(eigvals))
axs[1].set_title("Diagonalized")
axs[1].imshow(P.transpose() @ XXt @ P)
plt.show()
print(np.linalg.matrix_rank(X))
1.2872850938240181
100
T = 1000
eta = 0.001
ws = np.zeros((T, p, 1))
w = np.random.normal(size=(p, 1)) / np.sqrt(p)
losses = []
for t in range(T):
w = w - eta * XXt @ w + eta * XY
ws[t] = w
losses.append(np.linalg.norm(X.transpose() @ w - Y) ** 2 / n)
ws = ws.squeeze(axis=2)
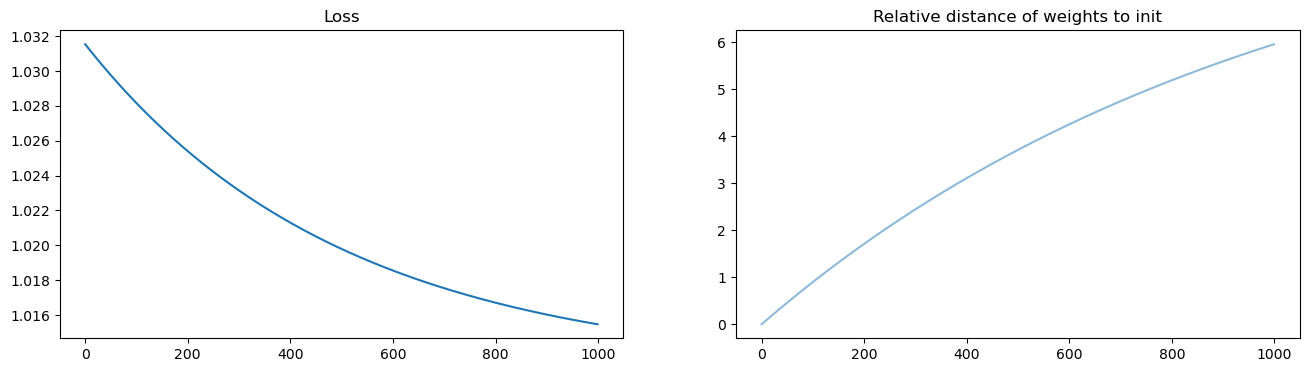
fig, axs = plt.subplots(1, 2, figsize=(16, 4))
axs[0].set_title("Loss")
axs[0].plot(losses)
axs[1].set_title("Relative distance of weights to init")
weight_evol = np.linalg.norm(ws - ws[0, :], axis=1) / np.linalg.norm(ws[0, :])
axs[1].plot(weight_evol, alpha=0.5)
plt.show()
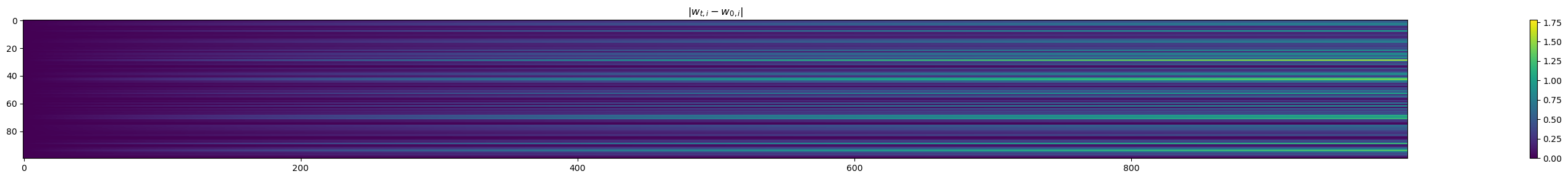
In the right basis, most coordinates of w dont move#
w only moves in a low dimensional subspace: the image of X
fig, ax = plt.subplots(figsize=(40, 3))
plt.title(r"$|w_{t, i} - w_{0, i}|$")
ws_in_base = ws @ P
a = ax.imshow(np.abs((ws_in_base - ws_in_base[0]).transpose()))
fig.colorbar(a)
plt.tight_layout()
plt.show()
Bias complexity tradeoff
xs = np.linspace(-1, 1, 100)
plt.plot(xs, simple_f(xs), label="Simple f")
plt.plot(xs, middle_f(xs), label="Middle f")
plt.plot(xs, complex_f(xs), label="Complicated f")
plt.title("Target functions")
plt.legend()
plt.show()
npoints = 50
dataset = Data(n=npoints, xmin=-1, xmax=1, noise_level=0.05, type="middle")
plot_data(dataset)
plt.legend()
plt.show()
class LinSin(nn.Module):
"""
A linear model with sinusoidal features. Assumes feature_dim is even.
"""
def __init__(self, feature_dim):
super().__init__()
self.feature_dim = feature_dim
self.lout = nn.Linear(self.feature_dim, 1)
self.weight_history = []
def phi(self, x):
"""
Computes $sin(2\pi k x), cos(2\pi k x) $ for k in $[feature_dim / 2]$
"""
r = 0.2 * np.pi * torch.arange(int(self.feature_dim / 2)) * x
return torch.concatenate([torch.sin(r), torch.cos(r)], axis=1)
def save_weights(self):
with torch.no_grad():
l = list(self.parameters())
self.weight_history.append(np.copy(l[-1].numpy()))
def forward(self, x):
return self.lout(self.phi(x))
model = LinSin(feature_dim=30)
losses = train(
model,
dataset,
N_steps=2000,
batch_size=npoints,
lr=0.01,
save_weights_every=-1,
)
plot_net(model)
plot_data(dataset)
plt.title("Trained linear model = dim 30")
plt.show()
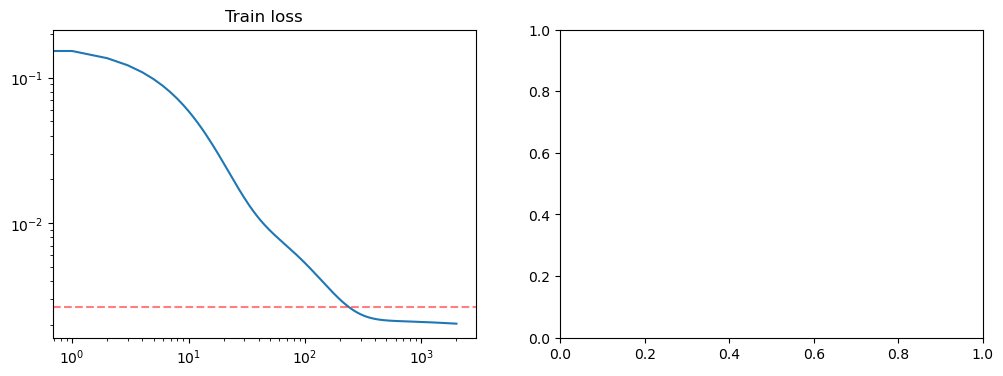
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
axs[0].loglog(losses)
axs[0].set_title("Train loss")
true_risk = dataset.risk(model)
print(true_risk)
axs[0].axhline(true_risk, color="red", alpha=0.5, linestyle="--")
plt.show()
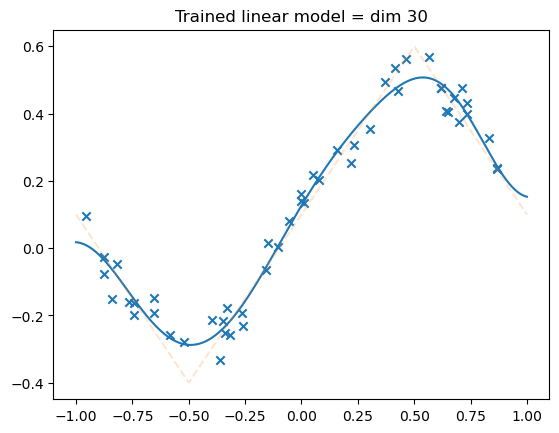
tensor(0.0026)
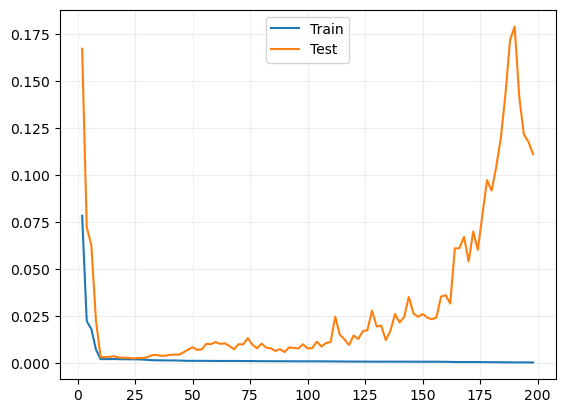
# train_losses = []
# var_train = []
# risks = []
dimensions = np.array(list(range(150, 200, 2))) # + list(range(100, 300, 2)))
for dim in dimensions:
losses = []
local_risks = []
for _ in range(10):
model = LinSin(feature_dim=dim)
losses.append(
train(
model,
dataset,
N_steps=1000,
batch_size=100,
lr=0.1,
save_weights_every=-1,
)[-1]
)
local_risks.append(dataset.risk(model).detach())
train_losses.append(np.mean(losses))
var_train.append(np.var(losses))
risks.append(np.mean(local_risks))
plt.grid(alpha=0.2)
plt.plot(np.arange(2, 200, 2), train_losses, label="Train")
plt.plot(np.arange(2, 200, 2), risks, label="Test")
plt.legend()
<matplotlib.legend.Legend at 0x29090d610>
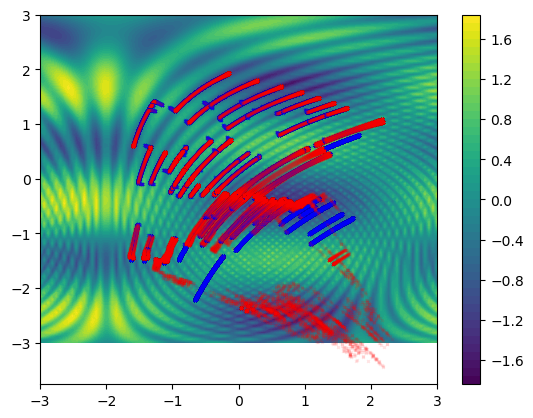
Gradient flow and Gradient descent
T = 10000
lrsmall = 0.0001
lrbig = 0.002
N = 100
with torch.no_grad():
xs = torch.linspace(-3, 3, 100)
ys = torch.linspace(-3, 3, 100)
xx, yy = torch.meshgrid(xs, ys, indexing="xy")
plt.contourf(xs, ys, f(xx, yy), levels=50)
plt.colorbar()
finalvals_small = []
finalvals_big = []
for i in range(N):
w0 = 3 * (torch.rand(2) - 0.5)
w0prime = w0.detach().clone()
wsmall = W(w0=w0)
wbig = W(w0=w0prime)
weights_small = []
weights_big = []
opt_small = optim.SGD(wsmall.parameters(), lr=lrsmall)
opt_big = optim.SGD(wbig.parameters(), lr=lrbig)
for _ in range(T):
opt_small.zero_grad()
wsmall().backward()
opt_small.step()
opt_big.zero_grad()
wbig().backward()
opt_big.step()
with torch.no_grad():
weights_small.append(np.copy(wsmall.param))
weights_big.append(np.copy(wbig.param))
finalvals_small.append(wsmall().detach())
finalvals_big.append(wbig().detach())
plt.plot(
[w[0] for w in weights_small],
[w[1] for w in weights_small],
"x",
alpha=0.1,
markersize=2,
label=i,
color="blue",
)
plt.plot(
[w[0] for w in weights_big],
[w[1] for w in weights_big],
"x",
alpha=0.1,
markersize=2,
label=i,
color="red",
)
plt.show()
plt.hist(finalvals_small, color="blue")
plt.show()
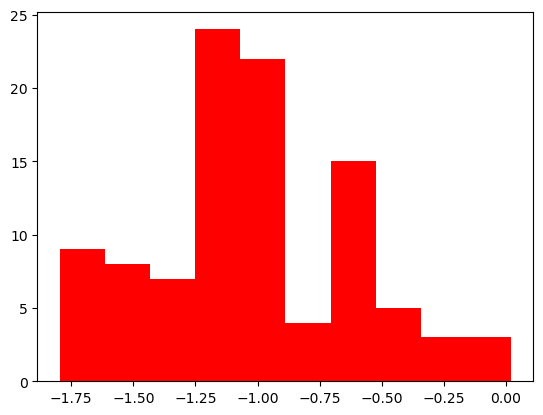
plt.hist(finalvals_big, color="red")
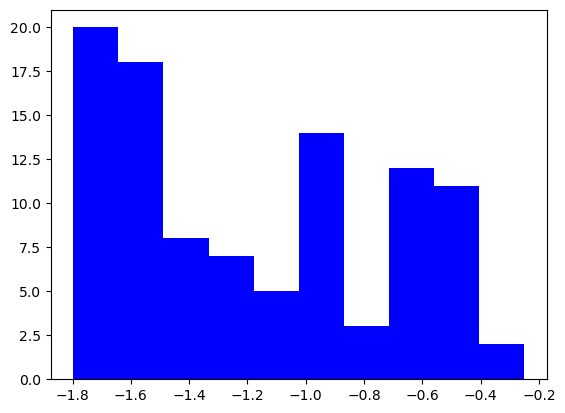
(array([ 9., 8., 7., 24., 22., 4., 15., 5., 3., 3.]),
array([-1.79505253, -1.61360693, -1.43216133, -1.25071573, -1.06927013,
-0.88782459, -0.706379 , -0.5249334 , -0.34348783, -0.16204225,
0.01940334]),
<BarContainer object of 10 artists>)
def f(x, y):
return torch.sin(x + 4) * (torch.cos(3 * y - 5) + 0.3) + 0.5 * torch.cos(
2 * (x + 2) * (y - 4) ** 2
)
class W(nn.Module):
def __init__(self, w0=None):
super().__init__()
if w0 is None:
w0 = 3 * (torch.rand(2) - 0.5)
self.param = nn.Parameter(w0)
def forward(self):
e0 = torch.tensor([1.0, 0.0])
e1 = torch.tensor([0.0, 1.0])
return f(e0 @ self.param, e1 @ self.param)